API#
-
class Aestivation#
- #include <Aestivation.h>
Implements aestivative behaviour for the model.
Only adult mated females aestivate, going into aestivation within a hiding time window determined by
t_hide1andt_hide2and emerging from aestivation within a waking time window determined byt_wake1andt_wake2. The females are isolated from their original patches during aestivation and returned back at the end, dependent on mortality. Other parameter rates controlling aestivation arepsiandmu_aes, the aestivation rate and aestivation mortality respectively.See also
Public Functions
-
Aestivation(AestivationParams *params, int sites_size)#
Aestivation constructor.
See also
- Parameters:
params – aestivation parameters
sites_size – number of sites in the model, size of Model::sites
-
void hide(std::vector<Patch*> &sites)#
Hides aestivating females from their patches.
The number of females that attempt to go into aestivation (of the given genotype combination) is determined by a binomial distribution of the aestivation rate. Of those, the number that survive is determined by a binomial distribution of 1 -
mu_aes(the aestivation mortality). Aestivating females are temporarily separated from the rest of the female patch population.See also
Note
Not all females survive aestivation. Only adult mated females aestivate.
- Parameters:
sites – [inout] vector of all Patch objects
-
void wake(int day, std::vector<Patch*> &sites)#
Wakes a fraction of the aestivating females.
The number of females that wake up on the given day (for the given genotype combination and patch) is determined by a binomial distribution with probability
1.0 / (1.0 + t_wake2 - (day%365). Those that wake are returned to their patch’s adult mated female population.See also
- Parameters:
day – [in] simulation day
sites – [inout] vector of all Patch objects
-
bool is_hide_time(int day)#
Determines if the time is within the aestivation hiding window.
t_hide1\( < t \le \)t_hide2, where \( t \) is the day of the current year. Assumespsi> 0.00001.Note
The hiding window is exclusive of the start time but inclusive of the end time.
- Parameters:
day – [in] simulation day
- Returns:
As you would expect.
-
bool is_wake_time(int day)#
Determines if the time is within the aestivation waking window.
t_wake1\( < t \le \)t_wake2, where \( t \) is the day of the current year. Assumespsi> 0.00001.Note
The waking window is exclusive of the start time but inclusive of the end time.
- Parameters:
day – [in] simulation day
- Returns:
As you would expect.
Private Members
-
double psi#
Aestivation rate.
-
double mu_aes#
Aestivation mortality.
-
int t_hide1#
Start day of aestivation-hiding period (exclusive).
-
int t_hide2#
End day of aestivation-hiding period (inclusive).
-
int t_wake1#
Start day of aestivation-waking period (exclusive).
-
int t_wake2#
End day of aestivation-waking period (inclusive).
-
Aestivation(AestivationParams *params, int sites_size)#
-
struct AestivationParams#
- #include <Params.h>
Aestivation model parameters.
Public Members
-
double psi#
Aestivation rate.
-
double mu_aes#
Aestivation mortality.
-
int t_hide1#
Start day of aestivation-hiding period (exclusive).
-
int t_hide2#
End day of aestivation-entering period (inclusive).
-
int t_wake1#
Start day of aestivation-waking period (exclusive).
-
int t_wake2#
End day of aestivation-waking period (inclusive).
-
double psi#
-
struct AreaParams#
- #include <Params.h>
Model area parameters.
Public Members
-
int num_pat#
Number of population sites chosen for the simulation.
-
int num_pat#
-
class BoundaryStrategy#
- #include <BoundaryStrategy.h>
Boundary strategy base class.
Defines the interface for boundary-type classes.
BoundaryStrategy classes implement methods specific to the boundary type of the model. The simulation area is assumed to have boundary conditions at x =
side_x, y =side_y, which are calculated from provided input coordinates or assumed to be 1 otherwise.Subclassed by EdgeBoundaryStrategy, ToroidalBoundaryStrategy
Public Functions
-
inline BoundaryStrategy(double side_x, double side_y)#
BoundaryStrategy constructor.
- Parameters:
side_x – [in] size of one side of the simulation area (x-axis)
side_y – [in] size of one side of the simulation area (y-axis)
-
inline virtual ~BoundaryStrategy()#
-
inline BoundaryStrategy(double side_x, double side_y)#
-
class Dispersal#
- #include <Dispersal.h>
Dispersal base class.
Defines the interface for dispersal classes.
The Dispersal classes compute the connections between Patch objects with their respective connection weights, dependent on the maximum dispersal distance
max_disp, and carry out dispersal in and out of the patches, dependent on the dispersal ratedisp_rate.See also
Subclassed by DistanceKernelDispersal, RadialDispersal
Public Functions
-
Dispersal(DispersalParams *params, BoundaryType boundary, double side_x, double side_y)#
Dispersal constructor.
Constructs the BoundaryStrategy object from the boundary type passed.
- Parameters:
params – [in] dispersal parameters
boundary – [in] boundary type to use for calculating dispersal distances
side_x – [in] size of one side of the simulation area (x-axis)
side_y – [in] size of one side of the simulation area (y-axis)
-
inline virtual ~Dispersal()#
Protected Functions
-
std::vector<std::array<long long int, constants::num_gen>> M_dispersing_out(const std::vector<Patch*> &sites)#
Determines the number of adult males (of each genotype) dispersing out from each patch.
The number of adult males of a given genotype dispersing from a given patch is determined by a random draw from a binomial distribution with probability of adult dispersal rate.
- Parameters:
sites – [in] vector of all Patch objects
- Returns:
The number of adult males dispersing out, divided by genotype and outgoing patch.
-
std::vector<std::array<std::array<long long int, constants::num_gen>, constants::num_gen>> F_dispersing_out(const std::vector<Patch*> &sites)#
Determines the number of adult mated females (of each genotype combination) dispersing out from each patch.
The number of adult females of a given genotype combination dispersing from a given patch is determined by a random draw from a binomial distribution with probability of adult dispersal rate.
- Parameters:
sites – [in] vector of all Patch objects
- Returns:
The number of adult mated females dispersing out, divided by female genotype, male sperm genotype and outgoing patch.
Protected Attributes
-
double disp_rate#
Adult dispersal rate.
-
double max_disp#
Maximum distance at which two sites are connected.
-
std::vector<std::vector<int>> connec_indices#
Connected patch indices ordered by each patch in Model::sites, such that the first element contains the indices of all the patches connected to the first sites patch, etc.
-
std::vector<std::vector<double>> connec_weights#
Connection weights of the connected patches ordered by each patch in Model::sites, such that the first element contains the connection weights between the first patch in sites and all the patches connected to it, etc.
-
BoundaryStrategy *boundary_strategy#
-
Dispersal(DispersalParams *params, BoundaryType boundary, double side_x, double side_y)#
-
class DistanceKernelDispersal : public Dispersal#
- #include <Dispersal.h>
Implements dispersive behaviour in the model for simple dispersal, where connection weights between patches are defined by a distance kernel.
All dispersing individuals are assumed to survive dispersal, and are guaranteed a connected patch to disperse to.
Public Functions
-
inline DistanceKernelDispersal(DispersalParams *params, BoundaryType boundary, double side_x, double side_y)#
DistanceKernelDispersal constructor.
- Parameters:
params – [in] dispersal parameters
boundary – [in] boundary type to use for calculating dispersal distances
side_x – [in] size of one side of the simulation area (x-axis)
side_y – [in] size of one side of the simulation area (y-axis)
-
~DistanceKernelDispersal()#
-
virtual void set_connecs(std::vector<Patch*> &sites) override#
Sets the inter-patch connectivities for dispersal.
If the distance between two patches is less than the the maximum dispersal distance
max_disp, they are deemed to be connected. The connection weight is determined by the difference between the maximum dispersal distance and the distance between those patches:weight = max_disp - distance
Note
Under this dispersal type, patches are deemed to be connected to themselves, resulting in self-dispersal. This is such that dispersal can take place even in 1-population models.
- Parameters:
sites – [in] vector of all Patch objects
-
virtual void adults_disperse(std::vector<Patch*> &sites) override#
Implements dispersal by adults from and to each patch, depending on the patch connectivities.
All dispersing individuals are assumed to survive dispersal, and are guaranteed a connected patch to disperse to. The number of males dispersing from a given patch to each of its connected patches is determined by a random draw from a multinomial distribution with probabilities equal to the connection weights. Similarly for the mated females.
See also
Note
Only adult males and mated females are assumed to disperse from the patches.
- Parameters:
sites – [inout] vector of all Patch objects
Private Functions
-
std::pair<std::vector<std::vector<int>>, std::vector<std::vector<double>>> compute_connecs(std::vector<Patch*> &sites)#
Computes the set of connection indices and weights for a group of patches.
If the distance between two patches is less than the the maximum dispersal distance
max_disp, they are deemed to be connected. The connection weight is determined by the difference between the maximum dispersal distance and the distance between those patches:weight = max_disp - distance
Note
Under this dispersal type, patches are deemed to be connected to themselves, resulting in self-dispersal. This is such that dispersal can take place even in 1-population models.
- Parameters:
sites – [in] vector of all Patches objects
- Returns:
The connections between all patches, divided into connection indices and connection weights. These are then organised in the same order as Model::get_sites(), where the first item represents all connections to the first Patch of the sites vector, etc.
-
inline DistanceKernelDispersal(DispersalParams *params, BoundaryType boundary, double side_x, double side_y)#
-
class EdgeBoundaryStrategy : public BoundaryStrategy#
- #include <BoundaryStrategy.h>
Implements methods for edge boundary conditions.
The simulation area is assumed to have edge boundary conditions at x =
side_x, y =side_y.Public Functions
-
inline EdgeBoundaryStrategy(double side_x, double side_y)#
-
inline ~EdgeBoundaryStrategy()#
-
inline EdgeBoundaryStrategy(double side_x, double side_y)#
-
class GDRelease#
- #include <GDRelease.h>
Gene drive release base class.
Defines the basic implementation of gene drive release in the model.
GDRelease classes implement the selection mechanism of the release sites from available patches and the release of gene drive mosquitoes into the Patch objects at the chosen release times. The number of drive heterozygous adult male mosquitoes released into each release site at each release time is determined by
num_driver_M.Subclassed by RandomGDRelease, SchedGDRelease
Public Functions
-
inline GDRelease(int num_driver_M, std::vector<int> rel_times)#
GDRelease constructor.
- Parameters:
num_driver_M – [in] number of drive heterozygous (WD) adult male mosquitoes per release
rel_times – [in] days on which the gene drive mosquitoes will be released
-
inline virtual ~GDRelease()#
Protected Functions
-
bool is_release_time(int day)#
Determines if the day is a chosen release time.
- Parameters:
day – [in] simulation day
- Returns:
As you would expect.
-
inline GDRelease(int num_driver_M, std::vector<int> rel_times)#
-
struct InitialPopsParams#
- #include <Params.h>
Initial population values for the model.
Public Members
-
int initial_WJ#
Number of initial juvenile mosquitoes with wild homozygous (WW) genotype for each age group.
-
int initial_WM#
Number of initial adult male mosquitoes with wild homozygous (WW) genotype.
-
int initial_WV#
Number of initial adult unmated female (virgin) mosquitoes with wild homozygous (WW) genotype.
-
int initial_WF#
Number of initial adult mated female mosquitoes with wild homozygous (WW) genotype.
-
int initial_WJ#
-
struct InputParams#
- #include <InputParams.h>
Input parameters for the simulation.
Public Members
-
int num_runs#
Number of simulation replicates to run.
-
int max_t#
Maximum simulated time (in days).
-
int num_pat#
Number of population sites chosen for the simulation.
-
double mu_j#
Juvenile density independent mortality rate per day.
-
double mu_a#
Adult mortality rate per day.
-
double beta#
Parameter that controls mating rate.
-
double theta#
Average egg laying rate of wildtype adult females (eggs per day).
-
double comp_power#
Parameter that controls the juvenile survival probability.
-
int min_dev#
Minimum development time for a juvenile (in days).
-
double gamma#
Rate of r2 allele formation from W/D meiosis.
-
double xi#
Somatic Cas9 expression fitness cost.
-
double e#
Homing rate in females.
-
int driver_start#
Time to start releasing drive alleles into the mosquito population.
-
int num_driver_M#
Number of drive heterozygous (WD) adult male mosquitoes per release.
-
int num_driver_sites#
Number of gene drive release sites per year.
-
double disp_rate#
Adult dispersal rate.
-
double max_disp#
Maximum dispersal distance at which two sites are connected.
-
double psi#
Aestivation rate.
-
double mu_aes#
Aestivation mortality rate.
-
int t_hide1#
Start day of aestivation-hiding period (exclusive).
-
int t_hide2#
End day of aestivation-hiding period (inclusive).
-
int t_wake1#
Start day of aestivation-waking period (exclusive).
-
int t_wake2#
End day of aestivation-waking period (inclusive).
-
double alpha0_mean#
Mean of the baseline contribution to the carrying capacity.
-
double alpha0_variance#
Variance of the baseline contribution to the carrying capacity.
-
double alpha1#
Rainfall contribution factor to carrying capacity.
-
double amp#
Amplitude of rainfall fluctuations.
-
double resp#
Carrying capacity’s responsiveness to rainfall contribution.
-
int initial_WJ = 10000#
Number of initial juvenile mosquitoes with wild homozygous (WW) genotype for each age group.
-
int initial_WM = 50000#
Number of initial adult male mosquitoes with wild homozygous (WW) genotype.
-
int initial_WV = 10000#
Number of initial adult unmated female (virgin) mosquitoes with wild homozygous (WW) genotype.
-
int initial_WF = 40000#
Number of initial adult mated female mosquitoes with wild homozygous (WW) genotype.
-
int rec_start#
Start time for the data recording window (inclusive).
-
int rec_end#
End time for the data recording window (inclusive).
-
int rec_interval_global#
Time interval for global data recording/output.
-
int rec_interval_local#
Time interval at which to collect/record local data (in days).
-
int rec_sites_freq#
Fraction of sites to collect local data for (1 is all sites, 10 is 1 in 10 etc).
-
int set_label#
‘Set of repetitions’ index label for output files.
-
int num_runs#
-
class InputRainfall : public Seasonality#
- #include <Seasonality.h>
Implements seasonality by modelling the rainfall contribution to carrying capacity from daily rainfall data.
Note
This class expects rainfall data for either every day of a year (365 days) or every day of the simulation (max_t).Annual data will be automatically looped for simulations longer than a year.
Public Functions
-
InputRainfall(InputRainfallParams *params)#
InputRainfall constructor.
- Parameters:
params – [in] seasonality parameters for input rainfall-type seasonality
-
inline ~InputRainfall()#
-
virtual double alpha(int day, double alpha0) override#
Computes the carrying-capacity alpha value for the given day and alpha0.
Models rainfall contribution as an exponential of daily rainfall.
\[ \alpha = \alpha_0 + \alpha_1 (1 - e^{-\textrm{R} r_d}), \]where \( \alpha \) is the carrying capacity, \( \alpha_0 \) is the baseline contribution, \( \alpha_1 \) is the factor accounting for rainfall contribution, \( \textrm{R} \) is the responsiveness to the rainfall contribution and \( r_d \) is the rainfall for the given day.- Parameters:
day – [in] simulation day
alpha0 – [in] baseline contribution to the carrying capacity
- Returns:
The carrying-capacity.
-
InputRainfall(InputRainfallParams *params)#
-
struct InputRainfallParams#
- #include <Params.h>
Seasonality model parameters for rainfall contribution to carrying capacity from rainfall data.
-
struct LifeParams#
- #include <Params.h>
Model life-process parameters.
Public Members
-
double mu_j#
Juvenile density independent mortality rate per day.
-
double mu_a#
Adult mortality rate per day.
-
double beta#
Parameter that controls mating rate.
-
double theta#
Average egg laying rate of wildtype adult females (eggs per day).
-
double comp_power#
Parameter that controls the juvenile survival probability.
-
int min_dev#
Minimum development time for a juvenile (in days).
-
double mu_j#
-
class Model#
- #include <Model.h>
Sets up and runs the model.
In addition to the set-up of the model, Model coordinates the different classes to run daily steps of mosquito life processes and dispersal as well as other events like gene drive release, aestivation and seasonality. It also contains methods to interface with the Record class.
See also
Public Functions
-
Model(ModelParams *params, const std::array<std::array<std::array<double, constants::num_gen>, constants::num_gen>, constants::num_gen> &inher_frac, SineRainfallParams *season, double a0_mean, double a0_var, std::vector<int> rel_sites = {}, BoundaryType boundary = BoundaryType::Toroid, DispersalType disp_type = DispersalType::DistanceKernel, std::vector<Point> coords = {})#
Model constructor.
Builds the components of the model for sinusoid rainfall-type seasonality.
- Parameters:
params – [in] model parameters
inher_frac – [in] inheritance fraction
season – [in] seasonality parameters for sinusoid rainfall
a0_mean – [in] alpha0 mean (mean of the baseline contribution to the carrying capacity)
a0_var – [in] alpha0 variance (variance of the baseline contribution to the carrying capacity)
rel_sites – [in] release sites (indices relative to the coords vector)
boundary – [in] boundary type
disp_type – [in] dispersal type
coords – [in] site coordinates vector
-
Model(ModelParams *params, const std::array<std::array<std::array<double, constants::num_gen>, constants::num_gen>, constants::num_gen> &inher_frac, InputRainfallParams *season, double a0_mean, double a0_var, std::vector<int> rel_sites = {}, BoundaryType boundary = BoundaryType::Toroid, DispersalType disp_type = DispersalType::DistanceKernel, std::vector<Point> coords = {})#
Model constructor.
Builds the components of the model for input rainfall-type seasonality.
- Parameters:
params – [in] model parameters
inher_frac – [in] inheritance fraction
season – [in] seasonality parameters for input rainfall data
a0_mean – [in] alpha0 mean (mean of the baseline contribution to the carrying capacity)
a0_var – [in] alpha0 variance (variance of the baseline contribution to the carrying capacity)
rel_sites – [in] release sites (indices relative to the coords vector)
boundary – [in] boundary type
disp_type – [in] dispersal type
coords – [in] site coordinates vector
-
void initiate()#
Sets up the model architecture.
Populates sites and sets dispersal connections.
-
void run(int day)#
Runs the daily step of the model.
The life processes do not run during the initialisation day (day 0). However, gene drive release can be carried out on day 0.
- Parameters:
day – [in] simulation day
-
long long int calculate_tot_J()#
Returns the total number of juveniles across all ages and genotypes and across all patches.
See also
- Returns:
The total number of juveniles in the model run.
-
long long int calculate_tot_M()#
Returns the total number of adult males across all genotypes and across all patches.
See also
- Returns:
The total number of adult males in the model run.
-
long long int calculate_tot_V()#
Returns the total number of adult virgin (unmated) females across all genotypes and across all patches.
See also
- Returns:
The total number of adult virgin (unmated) females in the model run.
-
long long int calculate_tot_F()#
Returns the total number of adult mated females across all female and male sperm genotypes and across all patches.
See also
- Returns:
The total number of adult mated females in the model run.
-
std::array<long long int, constants::num_gen> calculate_tot_F_gen()#
Returns the total number of adult mated females of each genotype across all patches.
See also
- Returns:
The total number of adult mated females in the model run, divided by female genotype.
-
std::vector<Patch*> get_sites() const#
Returns the sites vector.
The patches within the vector are ordered according to order of creation. If specific coordinates have been set for patches, the sites vector will follow the same order as the coordinates vector of the Model constructor.
- Returns:
The sites vector, containing all Patch objects.
-
int get_day() const#
Returns the current simulation day.
Simulation days run from 0 (initialisation day) up to
max_t(inclusive).- Returns:
The simulation day.
-
double get_alpha(double alpha0)#
Returns the carrying capacity for the given alpha0 (the baseline contribution to carrying capacity) and for the current day.
See also
- Parameters:
alpha0 – [in] baseline contribution to the carrying capacity
Private Functions
-
double alpha0()#
Returns a random value for the baseline contribution to the carrying capacity
alpha0.The random value is obtained as a random draw from a lognormal distribution with desired mean
alpha0_meanand desired variancealpha0_variance.- Returns:
A random draw of
alpha0.
-
void populate_sites()#
Populates all sites with a wild mosquito population of different types (age and sex).
-
void set_dev_duration_probs(int min_time, int max_time)#
Sets probabilities of juvenile eclosion for different age groups.
- Parameters:
min_time – [in] minimum development time (in days)
max_time – [in] maximum development time (in days)
-
void run_step(int day)#
Runs the daily mosquito life-processes for all sites, including dispersal and aestivation.
- Parameters:
day – [in] simulation day
-
void juv_get_older()#
Carries out juvenile aging across all patches.
See also
-
void adults_die()#
Carries out adult mortality across all patches.
See also
-
void virgins_mate()#
Carries out mating across all patches.
See also
-
void lay_eggs()#
Carries out egg-laying across all patches.
See also
-
void juv_eclose()#
Carries out juvenile eclosion across all patches.
See also
Private Members
-
std::vector<Patch*> sites#
Vector of all Patch objects.
Contains the population sites over the whole simulation area.
-
Aestivation *aestivation#
-
Seasonality *seasonality#
-
int day_sim#
Current day of the simulation.
-
int num_pat#
Number of population sites (patches) chosen for the simulation.
-
double side_x#
Size of one side of the simulation area (x-axis).
-
double side_y#
Size of one side of the simulation area (y-axis).
-
InitialPopsParams *initial_pops#
Initial population values - common for all Patches.
-
int min_dev#
Minimum development time for a juvenile (in days).
-
std::array<double, constants::max_dev + 1> dev_duration_probs#
Array of probabilities of juvenile development duration for a new juvenile (index indicates the number of days to develop or, equivalently, the age class the new juvenile starts at).
-
std::array<std::array<std::array<double, constants::num_gen>, constants::num_gen>, constants::num_gen> inher_fraction#
Inheritance fraction.
f_ijk is the fraction of genotype k offspring from mother with genotype i mated to father with genotype j.
-
double alpha0_mean#
Mean of the baseline contribution to the carrying capacity.
-
double alpha0_variance#
Variance of the baseline contribution to the carrying capacity.
-
Model(ModelParams *params, const std::array<std::array<std::array<double, constants::num_gen>, constants::num_gen>, constants::num_gen> &inher_frac, SineRainfallParams *season, double a0_mean, double a0_var, std::vector<int> rel_sites = {}, BoundaryType boundary = BoundaryType::Toroid, DispersalType disp_type = DispersalType::DistanceKernel, std::vector<Point> coords = {})#
-
struct ModelParams#
- #include <Params.h>
Model parameters.
Public Members
-
AreaParams *area#
-
InitialPopsParams *initial#
-
LifeParams *life#
-
AestivationParams *aes#
-
DispersalParams *disp#
-
ReleaseParams *rel#
-
AreaParams *area#
-
class Patch#
- #include <Patch.h>
Contains the information of a local mosquito population.
The population is divided into four types: juveniles (J), adult males (M), adult virgin (unmated) females (V) and adult mated females (F). These are then subdivided into six genotypes (in this order): WW, WD, DD, DR, WR and RR composed of a wild-type allele (W), a drive-type allele (D) and a resistant-type allele (R). Juveniles are also subdivided into age groups, ordered from oldest (0 days left to eclosion) to youngest (
max_dev- 1 days left), and do not have a specific sex. The population can carry out life-processes and interface with other classes to introduce gene drive mosquitoes into the Patch, carry out dispersal in and out of the Patch and aestivate.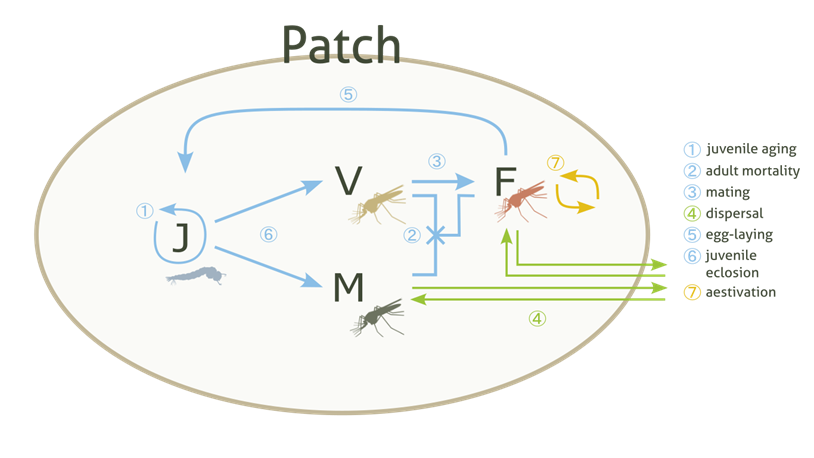
Public Functions
-
Patch(Model *mod, LifeParams *par, double a0, double side_x, double side_y)#
Patch constructor for randomly generated coordinates.
Sets coordinates randomly on the square simulation space of size side x side.
Note
Coordinates range from 0 to side for bound checking purposes.
- Parameters:
mod – [in] Model pointer
par – [in] life parameters
a0 – [in] alpha0 carrying capacity baseline
side_x – [in] size of one side of the simulation area (x-axis)
side_y – [in] size of one side of the simulation area (y-axis)
-
Patch(Model *mod, LifeParams *par, double a0, Point point)#
Patch constructor for custom coordinates.
- Parameters:
mod – [in] Model pointer
par – [in] life parameters
a0 – [in] alpha0 carrying capacity baseline
point – [in] patch coordinates
-
void populate(int initial_WJ, int initial_WM, int initial_WV, int initial_WF)#
Populates the local site with a wild mosquito population.
- Parameters:
initial_WJ – [in] The number of initial wild juveniles.
initial_WM – [in] The number of initial wild adult males.
initial_WV – [in] The number of initial wild adult virgin (unmated) females.
initial_WF – [in] The number of initial wild adult mated females.
-
std::array<long long int, constants::num_gen> get_M() const#
Returns the number of adult males in the patch, divided by genotype.
-
std::array<std::array<long long int, constants::num_gen>, constants::num_gen> get_F() const#
Returns the number of adult mated females in the patch, divided by female genotype and male sperm genotype.
-
std::array<long long int, constants::num_gen> get_F_fem_gen() const#
Returns the number of adult mated females in the patch, divided only by female genotype.
-
long long int calculate_tot_J()#
Calculates the total number of juveniles in the patch.
- Returns:
The total number of juveniles in the patch across all ages and genotypes.
-
long long int calculate_tot_M()#
Calculates the total number of adult males in the patch.
- Returns:
The total number of adult males in the patch across all genotypes.
-
long long int calculate_tot_V()#
Calculates the total number of adult virgin (unmated) females in the patch.
- Returns:
The total number of adult virgin (unmated) females in the patch across all genotypes.
-
long long int calculate_tot_F()#
Calculates the total number of adult mated females in the patch.
- Returns:
The total number of adult mated females in the patch across all female and male sperm genotypes.
-
void juv_get_older()#
Ages the juveniles of different age groups in the patch by a day.
The number of surviving individuals in an age group (for a given genotype) is determined by a binomial distibution of juvenile survival probability.
Note
Not all juveniles survive the aging process.
-
void adults_die()#
Removes dying adults from the patch.
Determines the number of adults that die in the given day and removes them from the patch. The number of adult males that die (for a given genotype) is determined by a binomial distribution of adult mortality, and similarly for adult virgin and mated females.
See also
-
void virgins_mate()#
Mates a fraction of the adult virgin females in the patch.
Determines the number of adult virgin females that mate in the given day (for a given female genotype) with a male of genotype j by using a binomial distribution of the mating rate. Then, tranforms the adult virgin females into adult mated females carrying male sperm of genotype j. Mating is carried out within the patch, and females only mate once.
-
void lay_eggs(const std::array<std::array<std::array<double, constants::num_gen>, constants::num_gen>, constants::num_gen> &f, const std::array<double, constants::max_dev + 1> &dev_duration_probs)#
Adult mated females lay eggs, creating new juveniles.
These juveniles do not have a specific sex.
Determines the number of eggs laid with genotype k on the given day by using a Poisson distribution. Other relevant parameters include the egg laying rate. Determines the development duration of these new juveniles using a multinomial distribution of development duration probabilities.
- Parameters:
inher_fraction – [in] inheritance fraction for new offspring
dev_duration_probs – [in] probabilities for juvenile development duration of new offspring
-
void juv_eclose()#
Turns the oldest juveniles into adults.
The number of survivors is determined by a binomial distribution of the juvenile survival probability. The proportion of the individuals’ sexes upon eclosion is determined by a binomial distribution with 0.5 probability.
Note
Not all juveniles survive eclosion.
-
void update_comp()#
Updates the juvenile survival probability of the patch.
Relevant parameters include the juvenile mortality rate, the juvenile survival probability power and the carrying capacity. The survival probability is computed as
\[ \textrm{comp} = (1 - \mu_j) \sqrt[\textrm{comp_power}]{\frac{\alpha}{\alpha + \textrm{J}_{\textrm{tot}}}}, \]with \( \mu_j \) juvenile mortality rate and \( \alpha \) carrying capacity.
-
void update_mate()#
Updates the mating rate of the patch.
Relevant parameters include the beta parameter. The mating rate is computed as
\[ \textrm{mate_rate} = \frac{\textrm{M}_{\textrm{tot}}}{\beta + \textrm{M}_{\textrm{tot}}}. \]See also
-
void M_disperse_out(const std::array<long long int, constants::num_gen> &m_out)#
Removes adult males from the patch as they disperse out.
- Parameters:
m_out – [in] The number of adult males dispersing out.
-
void F_disperse_out(const std::array<std::array<long long int, constants::num_gen>, constants::num_gen> &f_out)#
Removes adult mated females from the patch as they disperse out.
- Parameters:
f_out – [in] The number of adult mated females dispersing out.
-
void M_disperse_in(int gen, long long int m_in)#
Introduces new adult males into the patch as they disperse in.
- Parameters:
gen – [in] The genotype index for the adult males to disperse into M.
m_in – [in] The number of adult males dispersing in for the given genotype.
-
void F_disperse_in(int f_gen, int m_gen, long long int f_disp)#
Introduces new adult mated females into the patch as they disperse in.
- Parameters:
f_gen – [in] The female genotype index for the females to disperse into F.
m_gen – [in] The male sperm genotype index for the females to disperse into F.
f_disp – [in] The number of adult females dispersing in for the given genotype combination.
-
void F_hide(const std::array<std::array<long long int, constants::num_gen>, constants::num_gen> &f_try)#
Removes those females from the patch that attempt to go into aestivation.
- Parameters:
f_try – [in] The number of adult mated females that attempt to go into aestivation.
-
void F_wake(const std::array<std::array<long long int, constants::num_gen>, constants::num_gen> &f_wake)#
Introduces back into the patch those females that wake up from aestivation.
- Parameters:
f_wake – [in] The number of adult mated females from the given patch that wake up from aestivation.
-
void add_driver_M(int num_driver_M)#
Introduces driver heterozygous adult males into the patch.
- Parameters:
num_driver_M – [in] The number of driver heterozygous adult males to introduce.
Private Members
-
LifeParams *params#
Pointer to LifeParams object.
-
double alpha0#
Baseline contribution to the carrying capacity.
-
std::array<std::array<long long int, constants::max_dev + 1>, constants::num_gen> J#
Number of juvenile mosquitoes in the patch, divided by genotype and age group.
Note
The age groups are ordered from oldest (0 days left to eclosion) to youngest (max_dev - 1 days left). Juveniles do not have a specific sex.
-
std::array<long long int, constants::num_gen> M#
Number of adult male mosquitoes in the patch, divided by genotype.
-
std::array<long long int, constants::num_gen> V#
Number of adult virgin (unmated) female mosquitoes, divided by genotype.
-
std::array<std::array<long long int, constants::num_gen>, constants::num_gen> F#
Number of adult mated female mosquitoes F_{ij}, divided by female genotype i and male sperm genotype j.
-
long double comp#
Survival probability per juvenile per day (both density-dependent and independent factors).
-
long double mate_rate#
Probability of an adult virgin (unmated) female mating on a given day.
-
Patch(Model *mod, LifeParams *par, double a0, double side_x, double side_y)#
-
struct ProgressionParams#
- #include <Params.h>
Simulation progression parameters.
-
class RadialDispersal : public Dispersal#
- #include <Dispersal.h>
Implements dispersive behaviour for radial dispersal, where connection weights between patches are determined by the direction the patches are in relative to each other and other patches.
Particularly, the connection weight of a focal patch to its neighbouring patch is determined by the angle of bisecting lines from the centre of the focal patch to the catchment of the receiving patch. More distant villages may also be directly connected but the connectivity will be reduced if there are closer villages along the same flight path. The individuals that disperse in an unconnected direction will die.
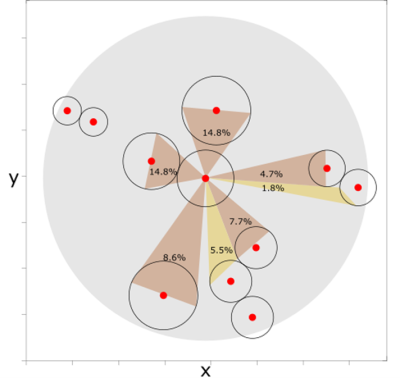
Public Functions
-
RadialDispersal(DispersalParams *params, BoundaryType boundary, double side_x, double side_y)#
RadialDispersal constructor.
- Parameters:
params – [in] dispersal parameters
boundary – [in] boundary type to use for calculating dispersal distances
side_x – [in] size of one side of the simulation area (x-axis)
side_y – [in] size of one side of the simulation area (y-axis)
-
~RadialDispersal()#
-
virtual void set_connecs(std::vector<Patch*> &sites) override#
Sets the inter-patch connectivities for radial dispersal.
If the distance between two patches is less than the the maximum dispersal distance, they may be connected. The connection weight of a focal patch to its neighbouring patch is determined by the angle of bisecting lines from the centre of the focal patch to the catchment of the receiving patch. More distant villages may also be directly connected but the connectivity will be reduced if there are closer villages along the same flight path. Patches that are further apart than the maximum dispersal distance are not connected.
- Parameters:
sites – [in] vector of all Patch objects
-
virtual void adults_disperse(std::vector<Patch*> &sites) override#
Implements dispersal by adults from and to each patch, depending on the patch connectivities.
Only those individuals that disperse out of a patch in a connected direction will survive. The number of dispersing males (of a given genotype) that survive dispersal out of their patch is determined by a random draw from a binomial distribution with probability of the total connection weight for all its connected patches. Of those, the number dispersing to each of the connected patches is determined by a random draw from a multinomial distribution with probabilities equal to the connection weights. Similarly for the mated females.
Note
Only adult males and mated females are assumed to disperse from the patches. There is also dispersal mortality associated with this dispersal type.
- Parameters:
sites – [inout] vector of all Patch objects
Private Functions
-
std::pair<std::vector<std::vector<int>>, std::vector<std::vector<double>>> compute_connecs(std::vector<Patch*> &sites)#
Computes the set of connection indices and weights for a group of patches.
If the distance between two patches is less than the the maximum dispersal distance, they may be connected. The connection weight of a focal patch to its neighbouring patch is determined by the angle of bisecting lines from the centre of the focal patch to the catchment of the receiving patch. More distant villages may also be directly connected but the connectivity will be reduced if there are closer villages along the same flight path. Patches that are further apart than the maximum dispersal distance are not connected.
Note
Under this dispersal type, patches are NOT connected to themselves.
- Parameters:
sites – [in] vector of all Patches objects
- Returns:
The connections between all patches, divided into connection indices and connection weights. These are then organised in the same order as Model::get_sites(), where the first item represents all connections to the first patch in Model::get_sites(), etc.
-
std::pair<std::vector<std::pair<double, double>>, double> compute_interval_union(const std::pair<double, double> &qq, const std::vector<std::pair<double, double>> &input)#
Computes the union of overlapping intervals and calculates the difference in the sum of lengths between the merged intervals and the original intervals.
This function takes a single interval
qqand a vector of intervalsinput. It merges any overlapping intervals, includes the non-overlapping intervals as they are, and calculates the total sum of lengths of the resulting intervals. It also computes the difference between the total sum of lengths of the merged intervals and the original interval. This is required to compute the extent of flight path between a focal patch and a recieving patch, while accounting for the shadowing effect of nearer patchesNote
The function assumes that each interval in the input is well-formed, meaning that for each interval
std::pair<double, double>, the first value (start) is less than or equal to the second value (end).- Parameters:
qq – [in] A pair of doubles representing the interval to be merged with the input intervals. The first value is the start of the interval, and the second value is the end of the interval.
input – [in] A vector of pairs of doubles, where each pair represents an interval. Each interval has a start value (first) and an end value (second).
- Returns:
A pair consisting of:
A vector of pairs of doubles representing the union of the merged and non-overlapping intervals, sorted by the start of each interval.
A double representing the difference between the sum of lengths of the merged intervals and the original intervals.
-
double wrap_around(double value, double range)#
Function to ‘wrap’ a real-valued number into the interval from zero to a maximum specified by the parameter ‘range’: if the input value is outside the interval it is wrapped around so that the output is within the interval.
- Parameters:
value – [in] the real-valued number to be wrapped around
range – [in] the maximum value of the interval to be wrapped into
- Returns:
the wrapped number within the interval
-
std::vector<int> get_sorted_positions(const std::vector<double> &numbers)#
Sorts the indices of the vector elements based on the numeric value of the corresponding element (in ascending order).
- Parameters:
numbers – [in] vector to sort
- Returns:
The sorted indices vector.
-
std::pair<std::vector<double>, std::vector<int>> compute_distances_site(int, std::vector<Patch*> &sites)#
Determines the points within a distance ‘max_dis’ of focal patch object from the set of all patch objects.
- Parameters:
i – [in] the index of the focal point
sites – [in] vector of all Patch objects
- Returns:
A list of the distances of these points, and a list of the indices within the set of all points
Private Members
-
std::vector<double> connec_weights_sum#
Sum of all the connection weights for each patch, ordered by Model::sites.
-
RadialDispersal(DispersalParams *params, BoundaryType boundary, double side_x, double side_y)#
-
class RandomGDRelease : public GDRelease#
- #include <GDRelease.h>
Implements gene drive release for randomised release sites.
Release sites are randomised upon each release time, with the number selected on each release dependent on
num_driver_sites.See also
Public Functions
-
RandomGDRelease(ReleaseParams *params)#
RandomGDRelease constructor.
- Parameters:
params – [in] gene drive release parameters
-
inline ~RandomGDRelease()#
Private Functions
-
virtual std::vector<Patch*> select_driver_sites(int day, const std::vector<Patch*> &sites) override#
Selects random release sites.
Relevant parameters include the number of driver sites.
See also
- Parameters:
day – [in] simulation day
sites – [in] vector of all Patch objects
- Returns:
The chosen release sites.
Private Members
-
int num_driver_sites#
Number of gene drive release sites per release.
-
RandomGDRelease(ReleaseParams *params)#
-
class Record#
- #include <Record.h>
Records model data.
Creates CoordinateList, Totals and LocalData output .txt files for each repetition of the model. CoordinateList: contains the coordinates of all recorded sites. Totals: contains the total numbers of adult males of each genotype across all sites, for each recorded day of the simulation. LocalData: contains the number of adult males of each genotype for each site on each simulated day that is recorded.
See also
Public Functions
-
Record(RecordParams *rec_params, int rep)#
Record constructor.
Creates LocalData, Totals and CoordinateList output .txt files.
Creates a subdirectory for output files in the current directory.
- Parameters:
rec_params – [in] recording parameters
rep – [in] initial repetition label for the given set of runs
-
~Record()#
Record destructor.
Resets the current filepath so the output_files directory can be found in the next set of runs.
-
void record_coords(const std::vector<Patch*> &sites)#
Records the coordinates of the population sites.
Relevant parameters include the fraction of sites to collect data for.
See also
- Parameters:
sites – [in] vector of all Patch objects
-
void record_global(int day, const std::array<long long int, constants::num_gen> &tot_M_gen)#
Records the total numbers of adult mated female mosquitoes for the given day, divided by female genotype.
The totals are assumed to be across all sites.
See also
- Parameters:
day – [in] simulation day
tot_fem_gen – [in] total number of adult mated females divided by female genotype
-
void output_totals(int day, long long int tot_J, long long int tot_M, long long int tot_V, long long int tot_F)#
Outputs the total numbers of juvenile (J), adult male (M), adult virgin female (V) and adult mated female (F) mosquitoes for the given day.
The totals are assumed to be across all sites, and over all genotypes and age groups.
See also
- Parameters:
day – [in] simulation day
tot_J – [in] total number of juveniles
tot_M – [in] total number of adult males
tot_V – [in] total number of adult virgin (unmated) females
tot_F – [in] total number of adult mated females
-
void record_local(int day, const std::vector<Patch*> &sites)#
Records the number of adult mated females at each site for the given day.
The number of adult mated females at each site is divided by female genotype. Relevant parameters include the fraction of sites to collect data for.
See also
- Parameters:
day – [in] simulation day
sites – [in] vector of all Patch objects
-
bool is_rec_local_time(int day)#
Determines if it is time to record local data.
Other relevant parameters include the local recording interval.
See also
Record::record_local(), InputParams::rec_start, InputParams::rec_end, InputParams::rec_interval_local
Note
The initialisation day (day 0) will always be recorded, and the recording window will be inclusive of the start and end times.
- Parameters:
day – [in] simulation day
- Returns:
As you would expect.
-
bool is_rec_global_time(int day)#
Determines if it is time to record global data.
The number of adult males at each site is divided by genotype.
See also
- Parameters:
day – [in] simulation day
- Returns:
As you would expect.
Private Members
-
int rec_start#
Start time for the data recording window (in days) (inclusive)
-
int rec_end#
End time for the data recording window (in days) (inclusive)
-
int rec_interval_global#
Time interval for global data recording (in days)
-
int rec_interval_local#
Time interval for local data recording (in days)
-
int rec_sites_freq#
Fraction of sites to collect local data for (1 is all sites, 10 is 1 in 10 etc.).
-
int set_label#
‘Set of repetitions’ index label for output files.
-
int rep_label#
‘Repetition’ index label for output files (within given set).
-
int next_local_day#
-
int next_global_day#
-
std::ostringstream os1#
Local data output filename.
-
std::ostringstream os2#
Global data output filename.
-
std::ostringstream os3#
Coordinate data output filename.
-
std::ofstream local_data#
Local data filestream object.
-
std::ofstream global_data#
Global data filestream object.
-
std::ofstream coord_list#
Coordinate data filestream object.
-
Record(RecordParams *rec_params, int rep)#
-
struct RecordParams#
- #include <Params.h>
Data-recording parameters.
Public Members
-
int rec_start#
Start time for the data recording window (in days) (inclusive).
-
int rec_end#
End time for the data recording window (in days) (inclusive).
-
int rec_interval_global#
Time interval for global data recording/output.
-
int rec_interval_local#
Time interval at which to collect/record local data (in days).
-
int rec_sites_freq#
Fraction of sites to collect local data for (1 is all sites, 10 is 1 in 10 etc).
-
int set_label#
‘Set of repetitions’ index label for output files.
-
int rec_start#
-
class SchedGDRelease : public GDRelease#
- #include <GDRelease.h>
Implements gene drive release for pre-selected release sites.
Release sites are kept the same for each release, using the same pre-selected sites from the available Patches.
Public Functions
-
SchedGDRelease(ReleaseParams *params, std::vector<int> rel_sites, std::vector<Patch*> &sites)#
SchedGDRelease constructor.
- Parameters:
params – [in] gene drive release parameters
rel_sites – [in] chosen release sites, indices relative to Model::get_sites()
sites – [in] vector of all Patch objects
-
inline ~SchedGDRelease()#
Private Functions
-
SchedGDRelease(ReleaseParams *params, std::vector<int> rel_sites, std::vector<Patch*> &sites)#
-
class Seasonality#
- #include <Seasonality.h>
Seasonality base class.
Defines the interface for seasonality classes.
Subclassed by InputRainfall, SineRainfall
Public Functions
-
inline Seasonality(double alpha1)#
Seasonality constructor.
- Parameters:
alpha1 – [in] carrying capacity factor accounting for rainfall contribution
-
inline virtual ~Seasonality()#
-
virtual double alpha(int day, double alpha0) = 0#
Protected Attributes
-
double alpha1#
Carrying capacity factor accounting for rainfall contribution.
-
inline Seasonality(double alpha1)#
-
class Simulation#
- #include <Simulation.h>
Sets up and controls the flow of the simulation.
Running set_inheritance() and run_reps() in this order is essential to run a simulation. The class also contains setters for advanced options, which should be run before run_reps(). The simulation runs a burn-in period of 1 year (365 days) implicitly at the start of each repetition before outputting any data. A 1 year burn-in allows consistency in annual data such as rainfall (seasonality) data and aestivation periods.
See also
Note
Advanced options that have been set apply to all repetitions (runs) of a single simulation.
Warning
Advanced options will typically give an error message if the setting process has been unsuccessful. If they are not re-set successfully before running the simulation the option will maintain its default behaviour.
Public Functions
-
Simulation(InputParams input)#
Simulation constructor.
See also
- Parameters:
input – [in] simulation parameters
-
~Simulation()#
Simulation destructor.
-
void set_coords(const std::filesystem::path &filepath)#
Sets the sites’ coordinates and the release sites from a file, unless any errors are thrown.
The file should be structured in three columns, with x and y coordinates and whether the site is a release site (y/n) respectively. Values should be delimited by white space, and site rows should be delimited by new lines.
Warning
Setting specific release sites will change the mode of gene drive release from random to scheduled. The specified release sites will be used for each of the releases (if using multiple release times). Mosquitoes will be released at all release sites for each release time.
- Parameters:
filepath – [in] coordinates filepath, can be a relative or absolute filepath, or a filename (only if the file is in the build directory)
-
void set_boundary_type(BoundaryType boundary)#
Sets the boundary type for the model.
See also
- Parameters:
boundary – [in] boundary type
-
void set_dispersal_type(DispersalType disp)#
Sets the dispersal type for the model.
See also
- Parameters:
disp – [in] dispersal type
-
void set_rainfall(const std::filesystem::path &filepath)#
Sets the daily rainfall values from a file, unless any errors are thrown.
These can be daily values for a year cycle (365 days), or daily values for all the simulated days (max_t). The values should be delimited by new lines.
See also
Note
This option will use the input parameters alpha1 and resp previously provided and ignore amp.
- Parameters:
filepath – [in] rainfall filename, can be a relative or absolute filepath, or a filename (only if the file is in the build directory)
-
void set_release_times(const std::filesystem::path &filepath)#
Sets the release times for gene drive release from a file, unless any errors are thrown.
Release times should be simulation day numbers within the maximum time of simulation. Values should be delimited by new lines.
See also
Note
When using this option, the input parameter driver_start previously entered will be ignored.
- Parameters:
filepath – [in] release times filename, can be a relative or absolute filepath, or a filename (only if the file is in the build directory)
-
void set_inheritance(InheritanceParams inher_params)#
Sets the values of the f_{ijk} inheritance fraction for the gene drive considering r2 non-functional resistance alleles.
f_{ijk} denotes the fraction of genotype k offspring from mother with genotype i mated to father with genotype j.
The order of elements in each matrix axis is the following genotype order: WW, WD, DD, WR, RR and DR, composed of wild-type (W), drive-type (D) and resistant-type (R) alleles.
See also
InheritanceParams, InputParams:gamma, InputParams:xi, InputParams::e
Note
Six genotypes are counted and not nine because WD and DW genotypes are counted together, and likewise for the other heterozygous genotypes.
Note
DD, RR and DR adult females are assumed to be sterile as they don’t possess one functional copy of the dsx gene and thus produce no offspring.
- Parameters:
inher_params – [in] inheritance parameters
-
void run_reps()#
Runs the simulation num_runs times, recording data in output files.
See also
Private Members
-
int num_runs#
Number of simulation replicates to run.
-
int max_t#
Maximum simulated time (in days).
-
ModelParams *model_params#
Model parameters.
-
RecordParams *rec_params#
Data-recording parameters.
-
SineRainfallParams *sine_rainfall_params#
-
InputRainfallParams *input_rainfall_params#
Sinusoid rainfall seasonality parameters.
-
double alpha0_mean#
Input data rainfall seasonality parameters.
Seasonality parameter.
-
double alpha0_variance#
Seasonality parameter.
-
std::vector<int> release_sites#
Indices relative to coords vector for the gene drive release sites.
-
BoundaryType boundary_type#
-
DispersalType disp_type#
-
Simulation(InputParams input)#
-
class SineRainfall : public Seasonality#
- #include <Seasonality.h>
Implements seasonality by modelling the rainfall contribution to carrying capacity as a sinusoid wave.
Most useful for theoretical applications.
Public Functions
-
SineRainfall(SineRainfallParams *params)#
SineRainfall constructor.
- Parameters:
params – [in] seasonality parameters for sine rainfall-type seasonality
-
inline ~SineRainfall()#
-
virtual double alpha(int day, double alpha0) override#
Computes the carrying-capacity alpha value for the given day and alpha0.
Models rainfall contribution as a sine wave.
\[ \alpha = \alpha_0 + \alpha_1 \left(1 + \textrm{A}\sin\left(\frac{2 \pi d}{365}\right)\right), \]where \( \alpha \) is the carrying capacity, \( \alpha_0 \) is the baseline contribution, \( \alpha_1 \) is the factor accounting for rainfall contribution, \( \textrm{A} \) is the amplitude of rainfall fluctuations and \( d \) is the given simulation day.- Parameters:
day – [in] simulation day
alpha0 – [in] baseline contribution to the carrying capacity
- Returns:
The carrying-capacity.
Private Members
-
double amp#
Amplitude of rainfall fluctuations.
-
SineRainfall(SineRainfallParams *params)#
-
struct SineRainfallParams#
- #include <Params.h>
Seasonality model parameters for sinusoid rainfall contribution to carrying capacity.
-
class ToroidalBoundaryStrategy : public BoundaryStrategy#
- #include <BoundaryStrategy.h>
Implements the methods for toroidal boundary conditions.
The simulation area is assumed to have periodic boundary conditions at x =
side_x, y =side_y.Public Functions
-
inline ToroidalBoundaryStrategy(double side_x, double side_y)#
-
inline ~ToroidalBoundaryStrategy()#
-
inline ToroidalBoundaryStrategy(double side_x, double side_y)#
-
namespace constants#
Namespace for constants used throughout the program.
Variables
-
const int max_dev = 20#
Juvenile development time (egg to adult) expressed as days left till eclosion (eclosion on day 0).
-
const int num_gen = 6#
The number of different genotypes in the mosquito population.
For 6 genotypes these represent WW, WD, DD, WR, RR and DR (in this order), where W is the wild type allele, D is the drive type allele and R is the non-functional resistance allele.
-
const double pi = 3.14159265#
Numerical constant PI (to 8 d.p.).
-
const int max_dev = 20#
- file Aestivation.h
- #include <vector>#include <array>#include “Params.h”#include “Patch.h”#include “constants.h”
- file BoundaryStrategy.h
- #include “Point.h”
- file constants.h
Enums
-
enum BoundaryType#
An enum for the allowed boundary type values in the model.
Values:
-
enumerator Toroid#
Toroidal (or periodic) boundary conditions.
See also
-
enumerator Edge#
Edge boundary conditions.
See also
-
enumerator Toroid#
Variables
-
const int max_dev = 20#
Juvenile development time (egg to adult) expressed as days left till eclosion (eclosion on day 0).
-
const int num_gen = 6#
The number of different genotypes in the mosquito population.
For 6 genotypes these represent WW, WD, DD, WR, RR and DR (in this order), where W is the wild type allele, D is the drive type allele and R is the non-functional resistance allele.
-
const double pi = 3.14159265#
Numerical constant PI (to 8 d.p.).
-
enum BoundaryType#
- file Dispersal.h
- #include <vector>#include <array>#include <utility>#include “constants.h”#include “Params.h”#include “Patch.h”#include “Point.h”#include “BoundaryStrategy.h”
- file InputParams.h
- file inputval.h
- #include <iostream>#include <fstream>#include <string>#include <limits>#include <sstream>
Functions
-
template<typename T>
bool check_bounds(const std::string &par_name, T value, T lower_bound, bool inclusive_lower = true, T upper_bound = std::numeric_limits<T>::max(), bool inclusive_upper = true)# Checks if a value falls within bounds.
- Template Parameters:
T – type of the checked parameter
- Parameters:
par_name – [in] name of the parameter value
value – [in] parameter value
lower_bound – [in] value of the lower bound
inclusive_lower – [in] whether the lower bound value is included in the bounds
upper_bound – [in] value of the upper bound
inclusive_upper – [in] whether the upper bound value is included in the bounds
- Returns:
Whether the value falls within bounds or not.
-
template<typename T>
bool check_interval(const std::string &start_name, const std::string &stop_name, T start, T stop)# Checks if two values constitute a valid interval.
- Template Parameters:
T – type of the interval values
- Parameters:
start_name – [in] name of the interval start parameter
stop_name – [in] name of the interval stop parameter
start – [in] start value of the interval
stop – [in] stop value of the interval
-
template<typename T>
bool file_read_and_validate_type(std::ifstream &file, T &par, const std::string &par_name, const std::string &par_type)# Reads the next value from a filestream and assigns it to the parameter variable if the types match.
- Template Parameters:
T – type of the parameter value
- Parameters:
file – [inout] filestream
par – [out] parameter variable
par_name – [in] parameter name (for error messages)
par_type – [in] parameter type (for error messages)
- Returns:
Whether the assignment process has been successful.
-
template<typename T>
bool read_and_validate_type(std::stringstream &linestream, T &par, const std::string &par_name, const std::string &par_type)# Reads the next value from a linestream and assigns it to the parameter variable if the types match.
- Template Parameters:
T – type of the parameter value
- Parameters:
linestream – [inout] linestream
par – [out] parameter variable
par_name – [in] parameter name (for error messages)
par_type – [in] parameter type (for error messages)
- Returns:
Whether the assignment process has been successful.
-
template<typename T>
- file Model.h
- #include <vector>#include <array>#include “constants.h”#include “Params.h”#include “Patch.h”#include “Dispersal.h”#include “Aestivation.h”#include “GDRelease.h”#include “Seasonality.h”#include “Point.h”
- file Params.h
- #include <vector>
- file Patch.h
- #include <array>#include “Patch.h”#include “constants.h”#include “Model.h”#include “Params.h”#include “Point.h”
- file Point.h
- file random.h
- #include <vector>#include <array>#include “constants.h”
Functions
-
double random_real()#
Returns a random floating-point number from a uniform real distribution of 0.0 to 1.0.
- Returns:
The random number.
-
int random_discrete(int a, int b)#
Returns a random integer number from a uniform discrete distribution of a to b.
- Parameters:
a – [in] lower value of the distribution range
b – [in] upper value of the distribution range
- Returns:
The random number.
-
long long int random_poisson(double lambda)#
Returns a random draw (non-negative integer) from the Poisson distribution with mean lambda (using normal distribution approximation when lambda > 30)
- Parameters:
lambda – [in] mean of the distribution
- Returns:
The random number.
-
long long int random_binomial(long long int n, double p)#
Returns a random draw (non-negative integer) from the Binomial distribution B(N,p).
Uses Normal and Poisson distribution approximations for large N.
- Parameters:
n – [in] number of trials
p – [in] success probability
- Returns:
The random number.
-
std::vector<long long int> random_multinomial(long long int n, const std::vector<double> &probs)#
Returns a vector of outcomes from a random draw of the Multinomial distribution with N trials where each trial has a vector of probabilities probs.
- Parameters:
n – [in] number of trials
probs – [in] vector of probabilities for each outcome
- Returns:
A vector of the number of successes for each outcome (in the same order as the probabilities).
-
std::vector<long long int> random_multinomial(long long int n, const std::array<long long int, constants::num_gen> &probs)#
Returns a vector of outcomes from a random draw of the Multinomial distribution with N trials where each trial has a vector of probabilities probs.
- Parameters:
n – [in] number of trials
probs – [in] array of probabilities for each outcome (each genotype).
- Returns:
A vector of the number of successes for each outcome (in the same order as the probabilities).
-
std::vector<long long int> random_multinomial(long long int n, const std::array<double, constants::max_dev + 1> &probs)#
Returns a vector of outcomes from a random draw of the Multinomial distribution with N trials where each trial has a vector of probabilities probs.
- Parameters:
n – [in] number of trials
probs – [in] array of probabilities for each outcome (each age group)
- Returns:
A vector of the number of successes for each outcome (in the same order as the probabilities).
-
double random_lognormal(double des_mean, double des_var)#
Returns a random draw (non-negative floating-point number) from a lognormal distribution with desired mean des_mean and desired variance des_var.
- Parameters:
des_mean – [in] desired mean
des_var – [in] desired variance
- Returns:
The random number.
-
double random_real()#
- file Record.h
- #include <sstream>#include <fstream>#include <vector>#include <array>#include “constants.h”#include “Patch.h”#include “Params.h”
- file Seasonality.h
- #include <vector>#include “Params.h”
- file sets.h
- #include “InputParams.h”
Variables
-
InputParams set1#
Parameter set 1 - default.
Includes gene drive and dispersal but no aestivation or seasonality.
-
InputParams set2#
Parameter set 2 - 1 population.
Includes gene drive and dispersal but no aestivation or seasonality.
-
InputParams set3#
Parameter set 3 - no dispersal.
No dispersal via disp_rate = 0. Includes gene drive but no aestivation or seasonality.
-
InputParams set4#
Parameter set 4 - full mixing.
Full dispersal mixing via disp_rate = 1. Includes gene drive too but no aestivation or seasonality.
-
InputParams set5#
Parameter set 5 - 1 day.
Includes gene drive and dispersal but no aestivation or seasonality.
-
InputParams set6#
Parameter set 6 - high fitness cost.
Includes gene drive and dispersal but no aestivation or seasonality.
-
InputParams set7#
Parameter set 7 - low aestivation.
Includes gene drive and dispersal too but no seasonality.
-
InputParams set8#
Parameter set 8 - high aestivation.
Includes gene drive and dispersal too but no seasonality.
-
InputParams set9#
Parameter set 9 - no gene drive.
No gene drive via num_driver_M = num_driver_sites = 0. Includes dispersal but no aestivation or seasonality.
-
InputParams set10#
Parameter set 10 - high gene drive.
High gene drive via high num_driver_sites, num_driver_sites = num_pat. Includes dispersal too but no aestivation or seasonality.
-
InputParams set11#
Parameter set 11 - 1 population, full dispersal.
Full dispersal via disp_rate = 1. Includes gene drive too but no aestivation or seasonality.
-
InputParams set12#
Parameter set 12 - low seasonality, default rainfall.
Default rainfall contribution to seasonality modelled as a sinusoid wave - see SineRainfall. Low seasonality determined by low amp parameter. Includes gene drive and dispersal too but no aestivation.
-
InputParams set13#
Parameter set 13 - high seasonality, default rainfall.
Default rainfall contribution to seasonality modelled as a sinusoid wave - see SineRainfall. High seasonality determined by high amp parameter. Includes gene drive and dispersal too but no aestivation.
-
InputParams set14#
Parameter set 14 - low seasonality, pre-defined rainfall.
Pre-defined rainfall contribution to seasonality modelled from rainfall data file - see InputRainfall. Low seasonality determined by a low alpha0_mean : alpha1 ratio. Includes gene drive and dispersal too but no aestivation.
-
InputParams set15#
Parameter set 15 - high seasonality, pre-defined rainfall.
Pre-defined rainfall contribution to seasonality modelled from rainfall data file - see InputRainfall. Low seasonality determined by a high alpha0_mean : alpha1 ratio. Includes gene drive and dispersal too but no aestivation.
-
InputParams set16#
Parameter set 16 - alpha0 variance.
Includes a non-zero alpha0_variance to vary alpha0, the carrying capacity baseline, across patches. Includes gene drive and dispersal but no aestivation or seasonality.
See also
-
InputParams set17#
Parameter set 17 - multiple release times.
Includes gene drive and dispersal but no aestivation or seasonality.
Warning
Same as set 1, made to be used with a multiple gene drive release times file.
-
InputParams set18#
Parameter set 18 - high juvenile survival probability.
High juvenile survival probability via high comp_power. Includes gene drive and dispersal but no aestivation or seasonality.
-
InputParams set1#
- file Simulation.h
- #include <array>#include <vector>#include <string>#include <filesystem>#include “constants.h”#include “Params.h”#include “InputParams.h”#include “Point.h”
- file Aestivation.cpp
- #include <array>#include <vector>#include “Aestivation.h”#include “random.h”#include “constants.h”
- file BoundaryStrategy.cpp
- #include <cmath>#include “BoundaryStrategy.h”
- file Dispersal.cpp
- #include <vector>#include <array>#include <cmath>#include <limits>#include <algorithm>#include <numeric>#include “Dispersal.h”#include “random.h”#include “constants.h”#include <iostream>
- file GDRelease.cpp
- #include <vector>#include <algorithm>#include “GDRelease.h”#include “Patch.h”#include “random.h”
- file main.cpp
- #include <iostream>#include <string>#include <fstream>#include <filesystem>#include <limits>#include <chrono>#include <vector>#include “Simulation.h”#include “Params.h”#include “inputval.h”#include “InputParams.h”#include “sets.h”
- file main2.cpp
- #include <iostream>#include <string>#include <filesystem>#include “Simulation.h”#include “inputval.h”#include “constants.h”#include “Params.h”
- file Model.cpp
- #include <vector>#include <cassert>#include <algorithm>#include “Model.h”#include “random.h”#include “constants.h”#include “Patch.h”#include “Dispersal.h”#include “GDRelease.h”#include “Aestivation.h”
- file Patch.cpp
- #include <cmath>#include <array>#include <vector>#include “Patch.h”#include “random.h”#include “constants.h”
- file random.cpp
- #include <vector>#include <array>#include <random>#include <cmath>#include <algorithm>#include “random.h”#include “constants.h”
Functions
- std::mt19937 twister (1)
-
double random_real()
Returns a random floating-point number from a uniform real distribution of 0.0 to 1.0.
- Returns:
The random number.
-
int random_discrete(int a, int b)
Returns a random integer number from a uniform discrete distribution of a to b.
- Parameters:
a – [in] lower value of the distribution range
b – [in] upper value of the distribution range
- Returns:
The random number.
-
long long int random_poisson(double lambda)
Returns a random draw (non-negative integer) from the Poisson distribution with mean lambda (using normal distribution approximation when lambda > 30)
- Parameters:
lambda – [in] mean of the distribution
- Returns:
The random number.
-
long long int random_binomial(long long int n, double p)
Returns a random draw (non-negative integer) from the Binomial distribution B(N,p).
Uses Normal and Poisson distribution approximations for large N.
- Parameters:
n – [in] number of trials
p – [in] success probability
- Returns:
The random number.
-
std::vector<long long int> random_multinomial(long long int n, const std::vector<double> &probs)
Returns a vector of outcomes from a random draw of the Multinomial distribution with N trials where each trial has a vector of probabilities probs.
- Parameters:
n – [in] number of trials
probs – [in] vector of probabilities for each outcome
- Returns:
A vector of the number of successes for each outcome (in the same order as the probabilities).
-
std::vector<long long int> random_multinomial(long long int n, const std::array<long long int, constants::num_gen> &probs)
Returns a vector of outcomes from a random draw of the Multinomial distribution with N trials where each trial has a vector of probabilities probs.
- Parameters:
n – [in] number of trials
probs – [in] array of probabilities for each outcome (each genotype).
- Returns:
A vector of the number of successes for each outcome (in the same order as the probabilities).
-
std::vector<long long int> random_multinomial(long long int n, const std::array<double, constants::max_dev + 1> &probs)
Returns a vector of outcomes from a random draw of the Multinomial distribution with N trials where each trial has a vector of probabilities probs.
- Parameters:
n – [in] number of trials
probs – [in] array of probabilities for each outcome (each age group)
- Returns:
A vector of the number of successes for each outcome (in the same order as the probabilities).
-
double random_lognormal(double des_mean, double des_var)
Returns a random draw (non-negative floating-point number) from a lognormal distribution with desired mean des_mean and desired variance des_var.
- Parameters:
des_mean – [in] desired mean
des_var – [in] desired variance
- Returns:
The random number.
Variables
-
std::random_device rd#
- file Record.cpp
- #include <filesystem>#include <iostream>#include <sstream>#include <fstream>#include <vector>#include <iomanip>#include “Record.h”
- file Seasonality.cpp
- #include <cmath>#include <vector>#include “Seasonality.h”#include “constants.h”
- file Simulation.cpp
- #include <array>#include <vector>#include <iostream>#include <filesystem>#include <fstream>#include <sstream>#include <string>#include “Simulation.h”#include “constants.h”#include “Model.h”#include “Record.h”#include “inputval.h”
- dir /github/workspace/includes
- dir /github/workspace/src